Low cost diagnosis of common lysosomal storage disorders
Diagnosis of lysosomal storage disorders (LSD) is often challenging due to overlapping clinical symptoms and costly chemicals. We are developing a sequencing based assay that can diagnose over 20 LSDs simultaneously.

Lysosomal storage disorders are a group of 70 diseases that are caused by certain improper or non-functional lysosome enzymes. Individually, these diseases are rare but collectivelly, they occur in 1 in every 5000 child and often fatal.
Lysosomes are the “cleaners” inside the cells whose function is to dispose and recycle the waste products generated by the cells. When the lysosomes don’t function, these waste products accumulate all over the body leading to development of clinical symptoms such as- large spleen and liver, cheery red spots in the eyes, loss of learnt abilities (eg. talking and walking) etc.
Diagnosing these patients can help in giving life-saving treatment and provide counselling to the parents in subsequent pregnancies.
However, the diagnosis rate of these diseases is poor- just 5%. This suggests that vast majority of the patients are unknown to the healthcare services and dont receive life-saving treatments.
The problem is further complicated since each LSD is caused by a defect in a specific enzyme and different LSDs can have similar clinical symptoms. Thus, testing for a specific enzyme is costly and prone to error. Hence, in some cases, the diagnostic odyssey can range from 6 months to 10 years. Improving the diagnosis rate is imperative.
Our research group led by Dr. Jayesh Sheth and Dr. Harsh Sheth, together with Dr. Madhvi Joshi at the Gujarat Biotech Research Centre (GBRC), are developing a novel sequencing approach called single molecule molecular inversion probes (smMIPs; Figure 1) that can search for mutations in genes associated with 23 common LSDs at the same time. These are- Gaucher, Niemann-Pick A, Niemann-Pick B, Niemann-Pick C, Morquio-A, Morquio-B, Pompe, Fabry, Tay-Sachs, GM1 Gangliosidosis, Maroteaux-Lamy, Hurler, Hunter, Sanphilippo-A, Sanphilippo-B, Sandhoff, Mucolipidosis-II alpha/beta, Mucolipidosis-III alpha/beta, NCL-2, Metachromatic leukodystrophy and Krabbe diseases.
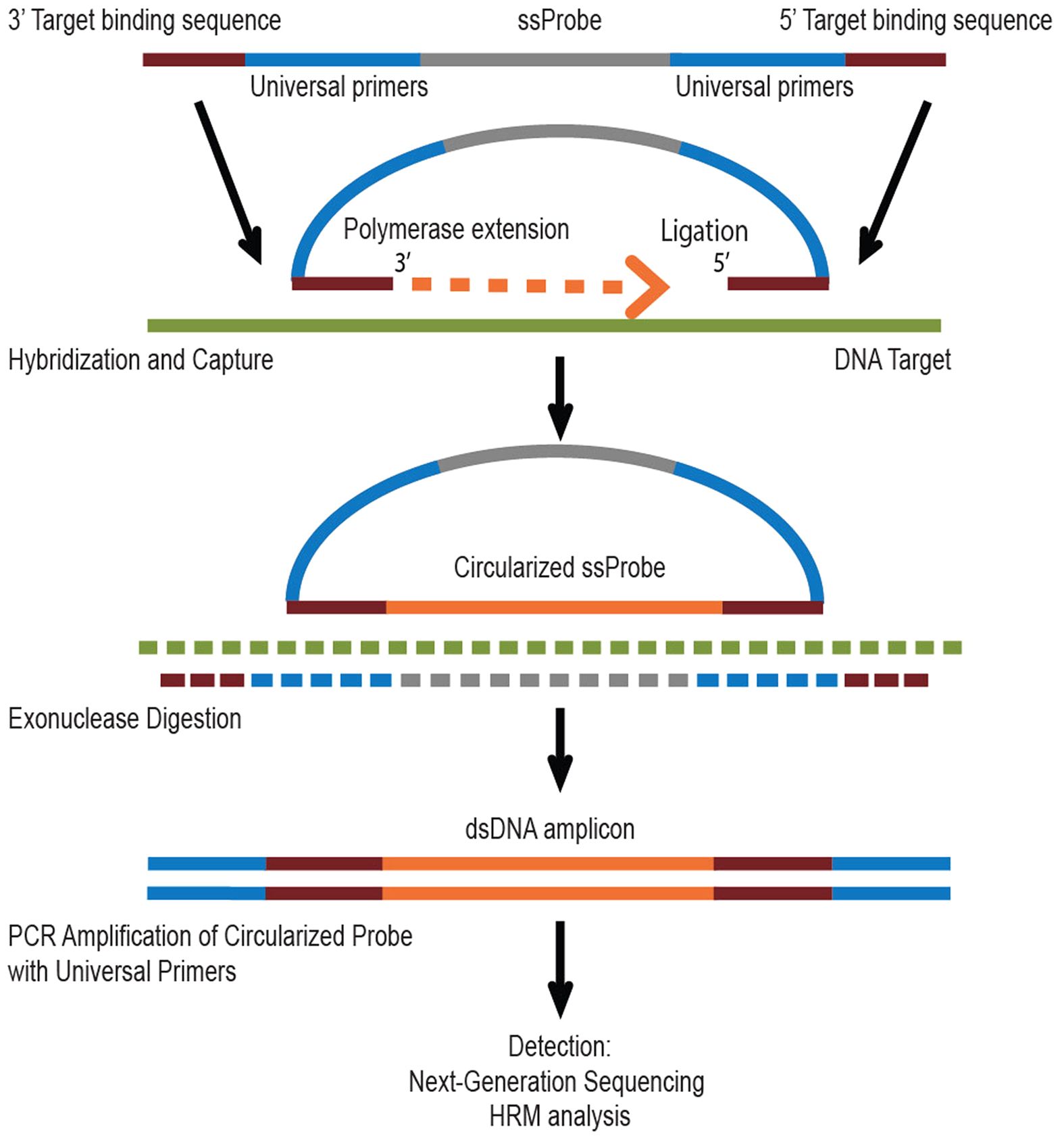
Figure 1: smMIP based target enrichment of specific DNA sequence. Figure taken from Stefan et al. Sci Rep 2016
The proposed approach is likely to improve diagnosis rate and reduce cost and time to diagnosis.
The work is funded by the Gujarat State Biotechnology Mission.